Challenges with Pancreatic Cancer
Pancreatic cancer is deemed the “silent killer” due to vague symptoms, late diagnosis, rapid progression, and poor response to therapy. As the 14th most common cancer but 7th leading cause of cancer-associated deaths, prolonging progression-free survival (PFS) and overall survival (OS) is a global public health priority.1 Current standard of care for pancreatic cancer is combination chemotherapy – FOLFIRINOX (FFX) or Gemcitabine + nab-paclitaxel (GnP).2
The COMPASS trial performed whole genome sequencing (WGS) and RNA sequencing (RNASeq) on pancreatic ductal adenocarcinoma (PDAC) patients prior to FFX or GnP first-line combination therapy.3 Early results from the COMPASS trial demonstrate that there are unique advanced PDAC genomic and transcriptomic subtypes with molecular heterogeneity between individuals and differing responses to chemotherapy. This is the first prospective evidence of the potential predictive power of molecular profiling.
NetraAI Advantage
NetraAI has unique capabilities from traditional machine learning (ML) efforts and can extract insights from multi-dimensional and highly heterogenous patient population datasets. By creating a mosaic of models or hypotheses, NetraAI can shed light on the genetic characteristics that are common to a specific subset of patients. These subsets are discovered from user provided labels. The NetraPlay system, an interactive causal AI playground enables users to interact with the subpopulations and explore different investigational scenarios to reveal underlying disease etiologies.
Using the COMPASS trial dataset of only 208 patients, NetraAI created NetraMaps focused on differentiating the causal factors that may be driving the patient response to FFX and GnP.
Key Findings
Genetic Mechanisms Differentiating FFX vs GnP Response in Stable Disease Patients
Using NetraAI to identify key drivers of differentiation between responses to FFX and GnP treatment in stable disease patients (between a 30% loss and 20% growth of tumor) revealed 3 distinct subpopulations (Figure 1).
The differentiation between Loop 1 and Loop 3 is driven by LRRC8E and PTPRH, downregulated and upregulated, respectively in Loop 1, and UTG1A13p which is highly expressed in Loop 3.
The NetraAI discovered genes driving patient subpopulations which are related to neuron death and regulation, neurotrophin signaling, and neuron apoptotic processes.
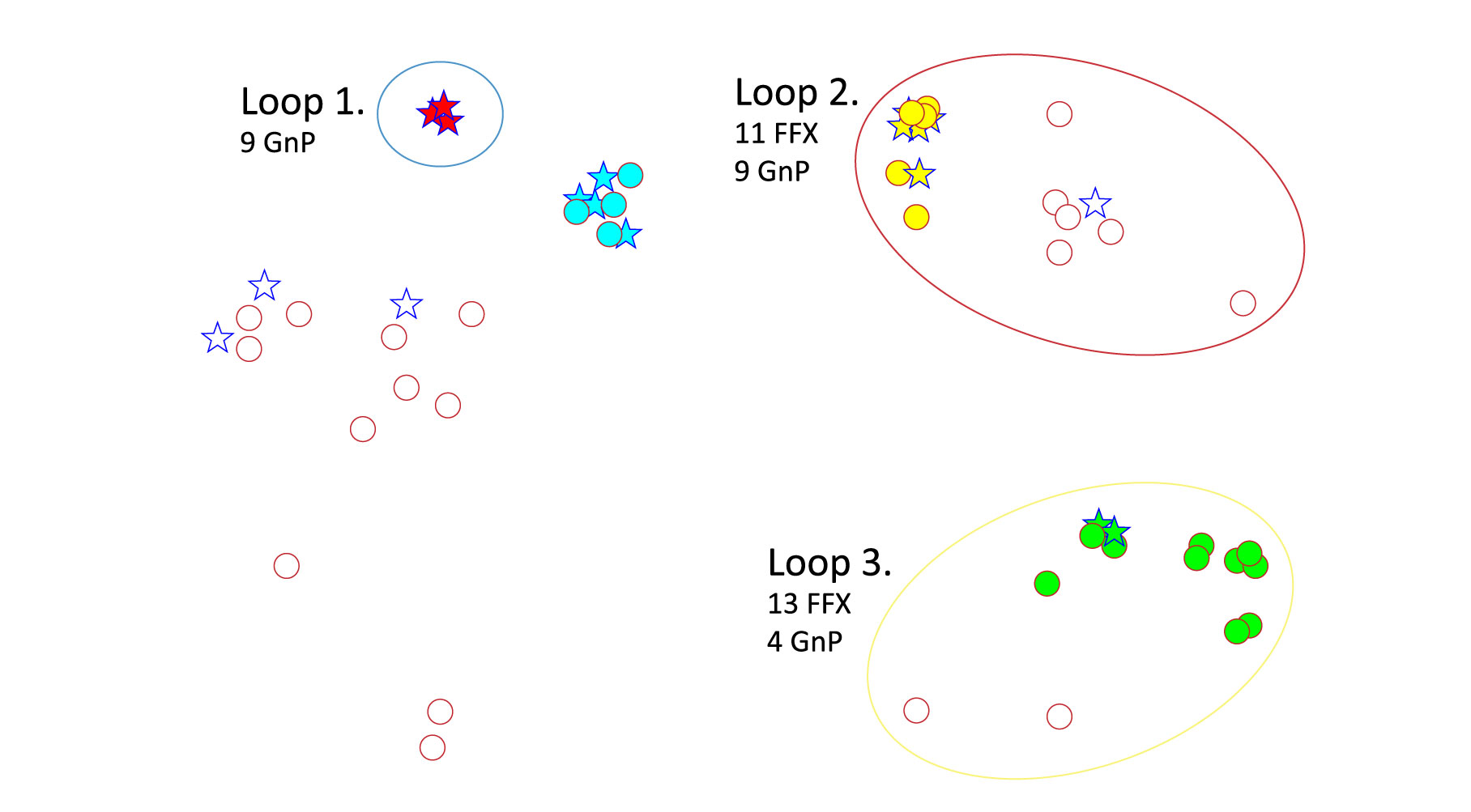
Blue Star = GnP Red Circle = FFX responder
Genetic Mechanisms Differentiating FFX vs GnP Response in Stable Disease, Partial Response, and Complete Response Patients
To extract more information from the patient dataset, we also had a map generated that identified drivers of differentiation between FFX and GnP patients with stable disease, partial response (between 100% and 30% loss of tumor), and complete response (complete disappearance of tumor) (Figure 2).
Loop 1 corresponds to 24 patients, 19 of which responded well to treatment with FFX, while Loop 2 corresponds to 84 patients, 44 who responded to FFX and 40 that responded well to GnP. The algorithm discovered this stratification because of a significantly higher expression of CLEC19A and LRRC29 in Loop 1 patients, suggesting that they may play a role in the mechanisms driving response to FFX and GnP. It is important to note that 78% of the individuals who responded well to GnP are contained within Loop 2.
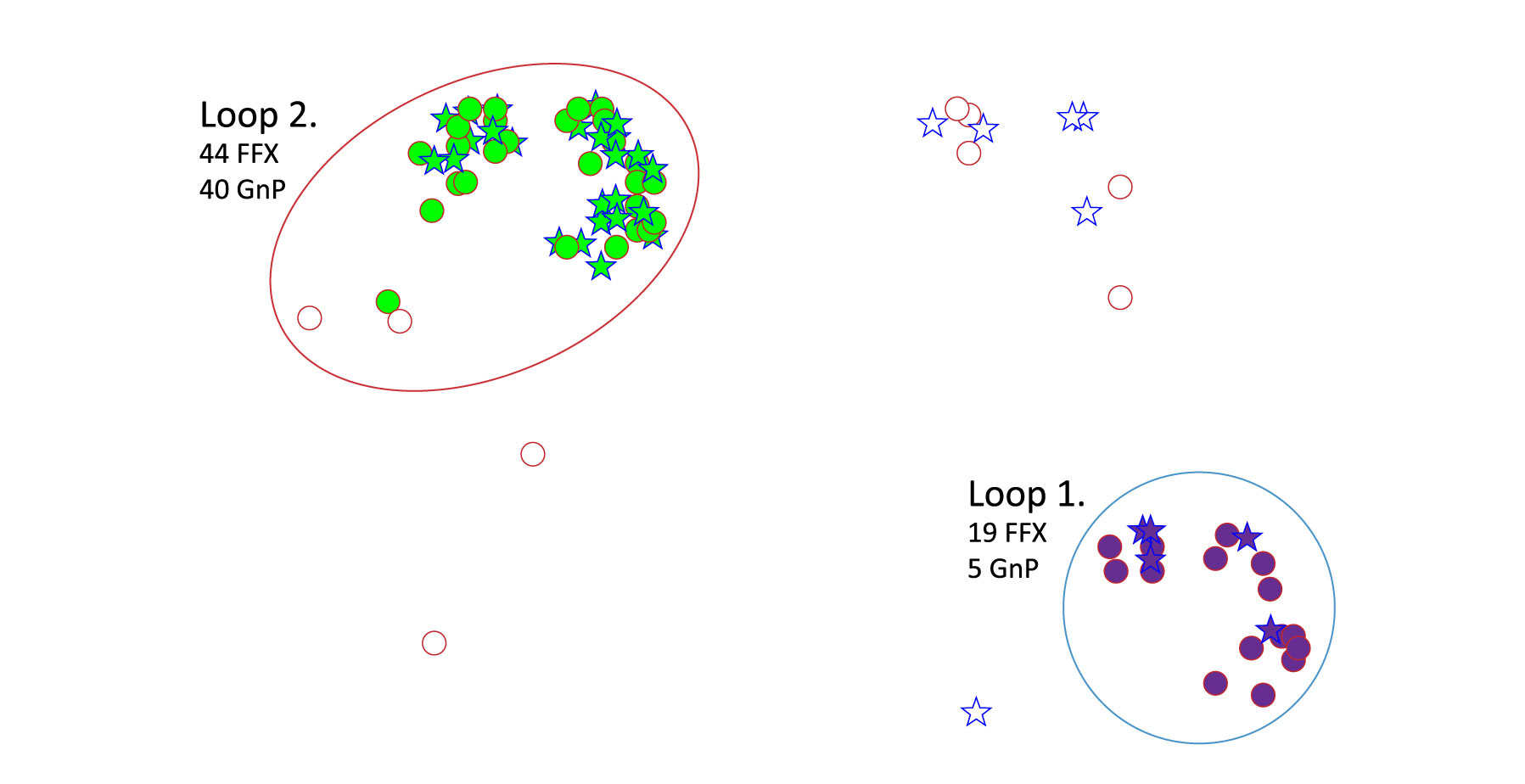
Figure 2. NetraAI identified PDAC subpopulation of FFX and GnP responders.
Blue Star = GnP Red Circle = FFX responder
Although neither are well understood, constructing a gene interaction network highlights that LRRC29 relates to HOOK1 (Figure 3). HOOK1 negatively regulates epithelial-to-mesenchymal transition by inhibiting SHP2 activity and has been found to be decreased in PDAC.4 Furthermore, the role of HOOK1 and SHP2 have been previously reported where the simultaneous decrease in expression levels are associated with a better patient response to GnP in nonsmall cell lung cancer.5 These results highlight that response to chemotherapy may be cancer agnostic and may depend on the chemotherapeutic agent and genetic factors involved in disease progression, with LRRC29 expression acting as a biomarker to choose between GnP and FFX treatment.
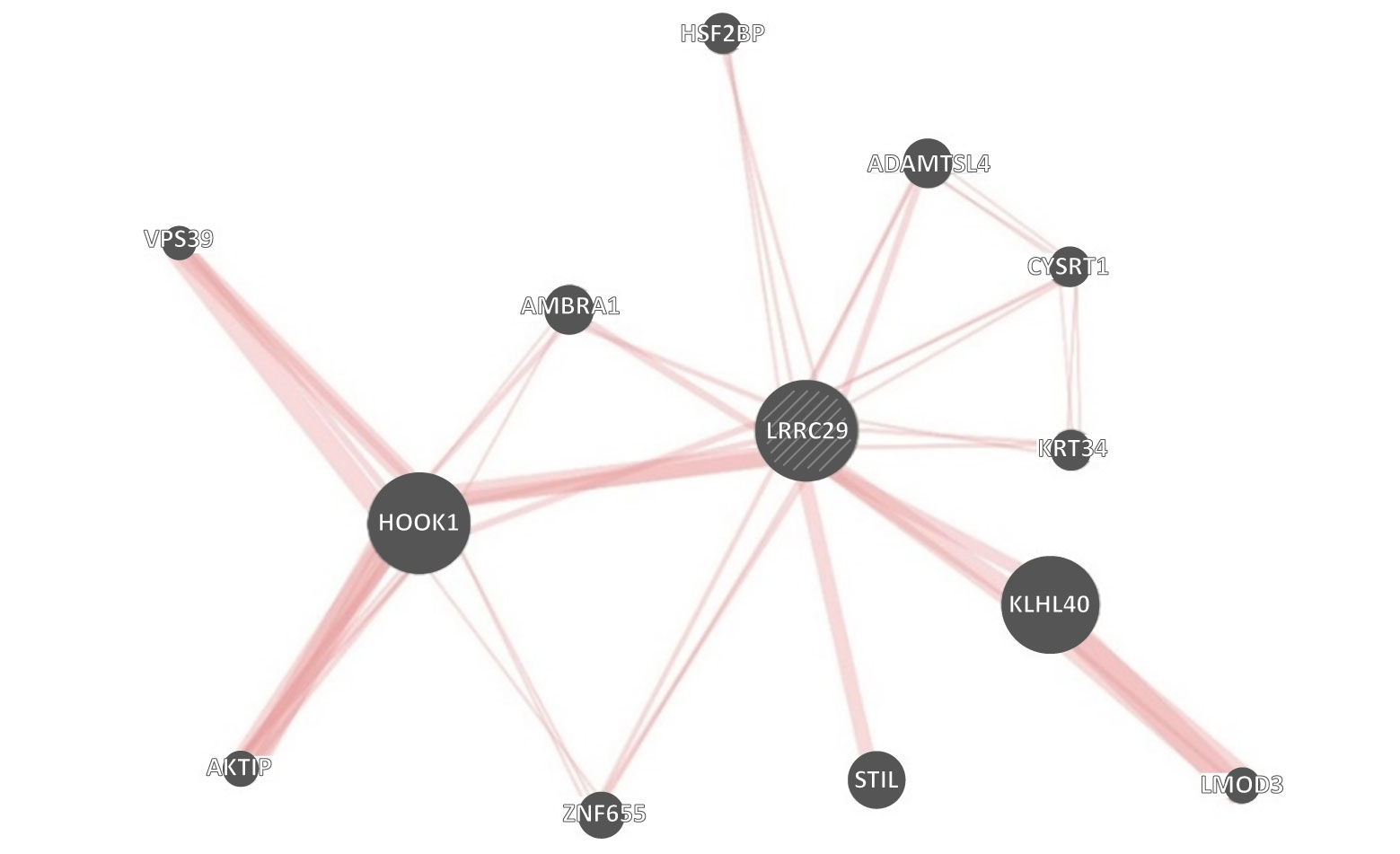
Red Lines = FFX responder
Significance of NetraAI and Impact on Precision Medicine
NetraAI has the unique capability to extract multiple hypotheses from patient datasets that allow us to draw conclusions on the mechanisms that play a role in specific patient subpopulations. This information can be used for biomarker discovery, targeted drug repurposing efforts, and additional areas of research focus such as identifying the underlying role of specific genes and pathways involved in regulating the patient response to therapy.
Using NetraAI, we were able to identify several distinct signatures within this small dataset alone that would allow oncologists to more confidently choose between FFX or GnP treatment for approximately 20% of patients. This research suggests that patients with decreased HOOK1 and SHP2 expression may better respond to GnP as previously reported;5 however, for PDAC patients with heightened LRRC29 expression, FFX may be the superior choice.
References
-
- Ushio, J. et al. Pancreatic Ductal Adenocarcinoma: Epidemiology and Risk Factors. Diagnostics 11, (2021).
- Klein-Brill, A., Amar-Farkash, S., Lawrence, G., Collisson, E. A. & Aran, D. Comparison of FOLFIRINOX vs Gemcitabine Plus Nab-Paclitaxel as First-Line Chemotherapy for Metastatic Pancreatic Ductal Adenocarcinoma. JAMA Netw Open 5, e2216199–e2216199 (2022).
- Aung, K. L. et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res 24, 1344–1354 (2018).
- Pan, Z. et al. Analysis of dynamic molecular networks for pancreatic ductal adenocarcinoma progression. Cancer Cell Int 18, 214 (2018).
- Yang, H. et al. The clinicopathological and prognostic implications of tyrosine phosphatase SHP2 and ankyrin Hook1 gene expression in non- small cell lung cancer patients treated with gemcitabine plus platinum as first-line chemotherapy. Ann Palliat Med 9, 2943952–2942952 (2020).

