Using NetraAI, driven by its novel Attractor AI-based system, we explored a gene expression dataset that included patients with OG with heterogenous outcomes. 1 We found a cohort of patients with differential gene expression patterns that were determined by transcriptomic variables related to human papillomavirus (HPV). An additional glioma methylation dataset was used with NetraAI yielded similar results with respect to HPV infection. 2 This demonstrates the ability of NetraAI to generate hypotheses that can be used by physicians to explore oncology patient subpopulations to discover novel drug targets, identify study confounds and understand heterogenous outcomes.
Glioma is a type of primary brain tumor originating from neural stem cells and/or glial progenitor cells with an accumulation of somatic mutations. Glioma includes several distinct subtypes that are related to the cell of origin and somatic mutations and/or translocations present; broadly, glioma includes glioblastoma (GBM), astrocytoma and oligodendroglioma (OG) as well as mixed pathologies.
NetraAI’s Role in Oligodendroglioma Research, Treatment, and Drug Discovery
NetraAI is a unique machine learning (ML) system that synergizes artificial intelligence (AI)-driven hypothesis generation with medical expertise to analyze complex multimodal data. Rooted in the Attractor AI paradigm, NetraAI excels at hypothesis generation to discover special combinations of variables through a unique lookback memory mechanism and extract valuable insights about patient subpopulations. A notable feature of NetraAI is that the technology works well with datasets with small numbers of patients and a high number of variables collected per patient as is typical in oncology. Notably, NetraAI has the potential to learn beyond the labels given, while also explaining the driving factors identified – in other words, it is optimized to find subpopulations of explainable patients that fit within the questions asked, e.g., drug response. These distinctive features allow for accurate and transparent definitions of patient subpopulations that provide a necessary tool for clinical trial success and risk mitigation. The intersection of human expertise and AI can revolutionize glioma research, uncovering explainable hypotheses for clinicians, researchers, and pharmaceutical companies that would be otherwise unattainable through traditional methods.
Unveiling the HPV Connection in Oligodendroglioma
Using a public OG dataset consisting of 156 primary OG tumors, 14 primary glioma samples, and 9 normal samples profiled using mRNA expression arrays, NetraAI was able to identify subpopulations of OG patients based on survival and differentially expressed genes. 1 A subpopulation of short-term survivors exhibited distinct HDAC1, LFNG, and RPS6KB1 gene expression. (Table 1)

Using functional genomic analysis, these genes and their functional pathways were found to have a common link to HPV infection through KEGG. Although HPV infection has been reported to have the potential to be considered an independent prognostic factor in GBM, there are no previous reports on a link to OG. 3 Most interestingly, NetraAI was able to very quickly highlight this association between OG and HPV infection status.
Further exploration revealed the HPV-related roles of HDAC1, LFNG, and RPS6KB1. In HPV-positive cervical cancer, the HPV E7 oncoprotein has been reported to interact with HDAC1, repressing several tumor suppressor genes. 4 Although LFNG is not directly related to the HPV pathway, it is involved in the Notch pathway which has been implicated in HPV-associated cancers as well as in gliomas, where NOTCH1 plays a role in glioma progression, contributing to the maintenance of glioma stem-like cells, cell proliferation, and resistance to apoptosis. 5 RPS6KB1 has been implicated in subtypes of HPV-positive cervical cancer, suggesting a role of the PI3K/Akt/mTOR pathway in HPV-positive cancers. 6
The NetraGPT module that uses the power of large language models (LLMs) provided the following output for these genes of interest, linking OG and HPV:
HDAC1 (Histone Deacetylase 1): involved in chromatin remodeling and gene expression regulation through histone deacetylation, which leads to a closed chromatin structure and transcriptional repression. It is vital role for cell cycle progression and differentiation.
Hypothesis: HDAC1 may contribute to OG pathogenesis by dysregulating gene expression critical for cell cycle control. HPV oncoproteins E6 and E7 may alter HDAC1 activity, promoting tumorigenesis by maintaining cells in a proliferative state.
LNFG (Lunatic Fringe): glycosyltransferase that modulates the Notch signaling pathway, which is crucial for cell differentiation, proliferation, and apoptosis. Aberrations in Notch signaling are associated with various cancers.
Hypothesis: LFNG may interact with HPV infection, potentially enhancing environments conducive to oncogenesis through dysregulated Notch signaling. LFNG mutations or altered expression could synergize with HPV infection to disrupt normal cell cycle checks, promoting OG development.
RPS6KB1 (Ribosomal Protein S6 Kinase B1): serine/threonine kinase involved in the PI3K/Akt/mTOR pathway, which regulates cell growth, survival, and metabolism. It is often overexpressed in various cancers and associated with aggressive tumor behavior.
Hypothesis: RPS6KB1 may facilitate OG progression when modulated by HPV infection. HPV oncoproteins may hijack the PI3K/Akt/mTOR signaling cascade through RPS6KB1 to promote uncontrolled cell growth and survival, providing a direct mechanistic link to tumor development and progression.
Integrative Hypothesis Combining All Elements
HPV could potentially contribute to the molecular etiology of OG by creating a permissive environment for mutations or by altering gene expression patterns directly through interactions with these critical regulatory pathways. This hypothesis provides a conceptual framework for exploring the potential oncogenic mechanisms driven by the combination of HPV infection and the alteration of specific genes in the development of OG. Further research would be needed to validate these hypothesized mechanisms.
Extrapolating the HPV-Glioma Link to Subpopulation Characterization Using Methylation Data
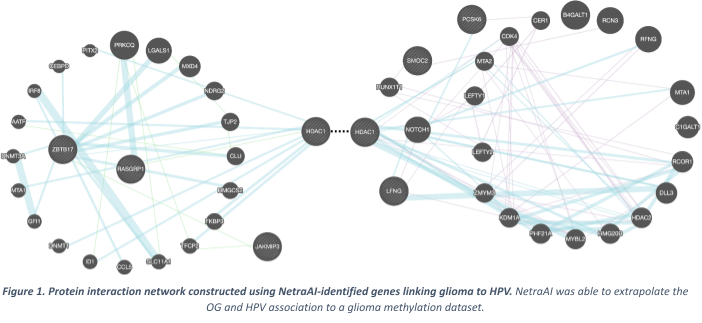
Prompted by the previous HPV-OG association, we extended our analysis to glioma methylation data using two reported datasets: one consisting of 293 lower-grade glioma patients7 and the other consisting of 1,122 pan-glioma (grade II-III-IV),2 and amalgamating a comprehensive dataset of 514 glioma patients with IDH mutation both with and without 1p/19q co-deletion. Using this dataset, NetraAI identified five differentially methylated genes – JAKMIP3, PCSK6, ZBTB17, RASGRP1, and SMOC2 – that are working in tandem in a subset of IDH-mutated patients. Using protein and gene interaction networks, we identify additional connections to HPV, that suggests a causal link between subtypes of glioma and HPV. Interestingly, these coordinated genes were also statistically significant when looked at as individual drivers.
Methylation of JAKMIP3 enables kinase and microtubule binding activity and is co-expressed with PPP1R13B. The HPV E7 oncoprotein has been reported to stimulated p53-mediated apoptosis through PPP1R13B. 8 Furthermore, JAKMIP3 is also co-expressed with CDKN2A, which has been reported to have a copy number loss in HPV-positive head and neck cancer patients, being associated with a poorer prognosis in terms of disease-free survival and overall survival. 9 These connections deepen the glioma-HPV connection and warrant further investigation.
With respect to PSCK6 methylation identified in a subpopulation of glioma patients, there is a protein interaction with GDF1, which has a direct gene interaction with MKI67. Additionally, ZBTB17, also has a direct gene interaction with MKI67 and has been reported to interact with the HPV E7 oncoprotein. 10 MKI67 encodes proliferation marker Ki-67 and elevated Ki-67 expression has been reported to have the worst prognosis in glioma patients. 11
RASGRP1, another differentially methylated marker in a subpopulation of glioma patients, was found to have direct gene interactions with ITLNI and FBXO31, which have a genetic and protein-protein interaction, respectively with CCNDI. CCNDI in turn, has been strongly implicated in HPV, where it has been found to be upregulated in HPV- head and neck cancers and downregulated in HPV+ head and neck cancers, suggesting that CCND1 downregulation may contribute to the survival of HPV+ head and neck cancer cells. 12
Finally, SMOC2 was found to be methylated in a subpopulation of glioma patients and has a direct gene interaction with ERBB2, which encodes HER2/Neu. In breast cancer, HPV genome was found most frequently in luminal A breast cancer tumors (ER+, PR+, HER2-) with low Ki67 expression. 13 This is in line with what was identified with NetraAI, where SMOC2 methylation was more significant in wildtype glioma patients and warrants further investigation in glioma pathogenesis and patient outcomes.
Evidently these 5 differentially methylated genes in subpopulations of glioma patients fortify the association between gliomas and HPV infection and warrant further investigation to elucidate mechanisms by which HPV infection plays a role in glioma pathogenesis to validate the discovery of potential therapeutic targets. A protein-protein interaction generated with these genes was created, demonstrating how they may cause alterations in the molecular machinery that drives certain subtypes of brain cancers. (Figure 1)
Significance of Findings and Application of NetraAI
NetraAI’s ability to generate a mosaic of etiological hypotheses allows for the quick exploration and consideration of potential targets and unveiled the potential impact of concomitant HPV infection on OG prognosis. Notably, this subpopulation stratification was not previously described by other groups investigating the same dataset, emphasizing the utility of the NetraAI approach in identifying subpopulations that may have important implications for the development of precision therapies for OG. NetraAI was able to provide novel insights into the molecular mechanisms underlying the OG-HPV connection, highlighting the potential role of HDAC1, LFNG, and RPS6KB1 as potential therapeutic OG targets, and subpopulation characterization. Insights derived from a glioma methylation dataset further solidified these findings as we were able to identify a set of 5 differentially methylated genes: JAKMIP3, PCSK6, ZBTB17, RASGRP1, and SMOC2 that simultaneously characterize subpopulations of glioma patients that further strengthen the HPV-glioma connection.
This research underscores the powerful synergy of Attractor AI and LLMs in dissecting patient population outcomes in clinical trials. By providing clear decompositions of patient populations into explainable and unexplainable subpopulations, we can derive generalizable insights that can help evaluate the chance of success of future trials from past trial data while simultaneously providing exclusion and inclusion criteria to enhance study endpoints and provide novel drug targets. Population-level investigations in the role of HPV on OG and glioma outcomes is therefore warranted. Practically, NetraAI’s ability to rapidly identify unknown subpopulations and provide etiological explanations is a testimony to its power in advancing clinical trials by empowering trialists with actionable exclusion/inclusion enrichment criteria for pivotal trials.
References
- Kamoun, A. et al. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat Commun 7, (2016).
- Ceccarelli, M. et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–563 (2016).
- Vidone, M. et al. Evidence of association of human papillomavirus with prognosis worsening in glioblastoma multiforme. Neuro Oncol 16, 298 (2014).
- Yeo-Teh, N. S. L., Ito, Y. & Jha, S. High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis. Int J Mol Sci 19, (2018).
- Bazzoni, R. & Bentivegna, A. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers (Basel) 11, (2019).
- Yang, S. et al. HPV‐related methylation‐based reclassification and risk stratification of cervical cancer. Mol Oncol 14, 2124 (2020).
- DJ, B. et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372, 2481–2498 (2015).
- Liebermann, D. A., Hoffman, B. & Vesely, D. Cell Cycle p53 Induced Growth Arrest versus Apoptosis and its Modulation by Survival Cytokines Extra View p53 Induced Growth Arrest versus Apoptosis and its Modulation by Survival Cytokines. Cell Cycle 6, (2007).
- Chen, W. S. et al. CDKN2A copy number loss in HPV- and HPV+ head and neck cancer to indicate poor prognosis: An integrated genomic and clinical TCGA analysis. https://doi.org/10.1200/JCO.2017.35.15_suppl.6060 35, 6060–6060 (2017).
- Morandell, D. et al. The human papillomavirus type 16 E7 oncoprotein targets Myc-interacting zinc-finger protein-1. Virology 422, 242 (2012).
- Zeng, A. et al. IDH1/2 mutation status combined with Ki-67 labeling index defines distinct prognostic groups in glioma. Oncotarget 6, 30232 (2015).
- Novotný, J. et al. Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas and Paired Normal Mucosae Reveals Cyclin D1 Deregulation and Compensatory Effect of Cyclin D2. Cancers (Basel) 12, 792 (2020).
13. Fernandes, A., Bianchi, G., Feltri, A. P., Pérez, M. & Correnti, M. Presence of human papillomavirus in breast cancer and its association with prognostic factors. Ecancermedicalscience 9, (2015)

